R Camp | day 1
Good morning, Detective
We’ve got a new case to crack. Let’s open RStudio.

Welcome to R Camp!
We are Kristie, Dorian, and Derek and we like R.
We are cats and not computer scientists.
We make lots of mistakes. You will see us make mistakes. Feel free to laugh at us. It’s okay.
Disclaimer
Spelling is very important in R.

1 | New project
Okay detective, let’s open a new dossier for today’s case.
Start a new project
- In Rstudio select File from the top menu bar
- Choose New Project…
- Choose New Directory
- Choose New Project
- Enter a project name such as
"cat_mystery" - Select Browse… and choose a folder where you normally perform your work.
- Click Create Project
1.1 New R script
R “scripts” are where you write your R code and document your work. They’re like recipes or prescriptions you write the tell the computer what you want to happen to your data.
Open a new R script
- In the upper left, click on the white file icon with the green (+) sign.
- Select R Script.

Save script
- Click on the floppy disk icon
- Enter a file name such as
cat_code.R
1.2 A tour of R Studio
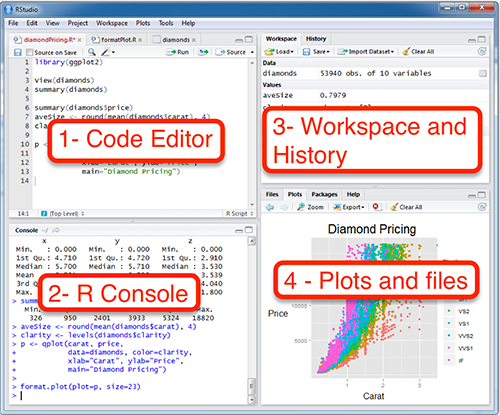
1. Code editor
This is where you write your scripts and document your work. The tabs at the top of the code editor allow you to view scripts and data sets you have open. This is where you’ll spend most of your time.
2. Console
This is where code is actually executed by the computer. It shows code that you have run and any errors, warnings, or other messages resulting from that code. You can input code directly into the console and run it, but it won’t be saved for later. That’s why we like to run all of our code directly from a script in the code editor.
3. Workspace
This pane shows all of the objects and functions that you have created, as well as a history of the code you have run during your current session. The environment tab shows all of your objects and functions. The history tab shows the code you have run. Note the broom icon below the Connections tab. This cleans shop and allows you to clear all of the objects in your workspace.
4. Plots and files
These tabs allows you to view and open files in your current directory, view plots and other visual objects like maps, view your installed packages and their functions, and access the help window. If at anytime you’re unsure what a function or package does, enter the name of thing after a question mark. For example, try entering ?mean into the console and push ENTER.
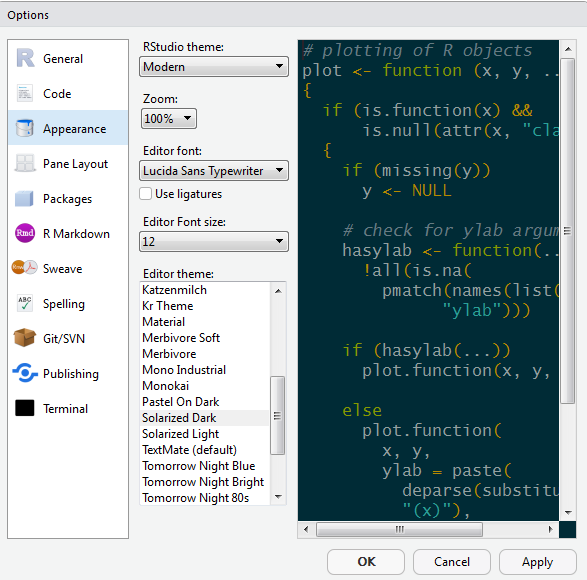
1.3 Make it your own
Let’s add a little style so we feel more at home. Follow these steps to change the font-size and and color scheme:
- Go to Tools on the top navigation bar.
- Choose
Global Options... - Choose
Appearancewith the paint bucket. - Find something you like.
2 | First steps
You can create objects and assign values to them using the “left arrow” <-, more officially known as the assignment operator. Try adding the code below to your R script and creating an object called name.
Once you add the code to your script, you can run the code by moving the blinking cursor to that line and pressing CTRL + ENTER.
# Create a new object
cat <- "Derek"
cat
# When saving text to a character object you need quotation marks.
# This won't work.
cat <- Demitri
# Without quotes, R looks for an object called Derek, and then let you know that it couldn't find one.
# You can copy an object by saving it to a new name.
cat2 <- cat
# Overwrite an object
cat <- "Demitri III"
cat
# Did name2 change as well?
cat2 If you create a name you don’t like you can drop it with the function rm().
# Delete objects to clean-up your environment
rm(cat)
rm(cat2)
# How can you get the original 'cat' object back?HOORAY! Don’t worry about deleting data or making a mistake in R. When you load data files into R it only copies the contents. That means all your original data files will remain safe and won’t suffer from any accidental changes. If anything disappears or goes wrong in R, it’s okay! You can always re-load the data using your script. No more worries about whether you remembered to save the latest data file or not.
2.1 Give it a name
Everything has a name in R and you can name things almost anything you like. You can even name your data TOP_SECRET_Shhhhhh... or unicorn_sightings or the_worst_data_ever.
Sadly, there are a few minor restrictions. Object names can’t include spaces or special characters such as a +, -, *, \, /, =, !, or ). However, on the plus side object names can include a _.
Your turn! Try running some of these examples in your R console.
n cats <- 5
n*cats <- 5
n_cats <- 5
n.cats <- 5
all_the_cats! <- "A very a big number"
# You can add one cat
n_cats <- n_cats + 1
# But what if you have 10,000 cats?
n_cats <- 10,000
# They also cannot begin with a number.
1st_cat <- "Fluffer Puff"
# But they can contain numbers.
cat1 <- "Fluffer Puff"NOTE: What happened when you created
n_catsthe second time?When you create a new object that has the same name as something that already exists, the new object will replace the old one. Sometimes you’ll want to update an existing object and replace the old version. Other times you may want to copy an object to a new name to preserve the original. This is similar to choosing between Save and Save As when you save a file.
2.2 Multiple items
You can put multiple values inside c() to make a vector of items. Each additional item is separated by a comma. The c stands for to conCATenate or to combine values.
Let’s use c() to create a few vectors of cat names and their ages.
# Create a character vector and name it cat_names
cat_names <- c("Fluffy", "Longmire", "Lucy")
# Print cat_names to the console
cat_names## [1] "Fluffy" "Longmire" "Lucy"# Create a numeric vector and name it cat_ages
cat_ages <- c(3,7,14.5)
# Print cat_ages to the console
cat_ages## [1] 3.0 7.0 14.52.3 Make a table
A table in R is known as a data frame. Data frames have columns of data, each made from a named vector. Let’s make a data frame with two columns by using the cat names and ages from above.
# Create table with columns "names" and "ages" with values from the cat_names and cat_ages vectors
cat_df <- data.frame(names = cat_names,
ages = cat_ages)
# Print the cat_df data frame to the console
cat_df## names ages
## 1 Fluffy 3.0
## 2 Longmire 7.0
## 3 Lucy 14.5To see the values in one of your columns, use the $ sign after the name of your table.
# Print the "ages" column in cat_df
cat_df$ages## [1] 3.0 7.0 14.5Pop Quiz, hotshot!
Which of these names are valid for a new object? (Hint: You are allowed to test them.)
my cat fred
my_CAT55
5cats
my-cat
Whatever!
Show solution
my_CAT55
Yes!! That was purrfect!
2.4 Leave a #comment
You may have noticed the text in the scripts with the # in front. These are called comments. Any line that starts with a # won’t be executed as R code. You can use the # to add notes in your script to make it easier for others and yourself to understand what is happening and why. You can also use comments to add warnings or instructions for others, add references to data you’re using, or point out things that need to be looked into further.
2.5 Functions

Now that you know what objects are and how to create them, let’s learn how to use them. Functions take one or more inputs called “arguments”. They perform steps based on the arguments and usually return an output object.
You can think of a function like ordering pizza.
order_pizza(address = "140 3rd place, Bingo MN",
toppings = c("mouse whiskers", "catnip", "anchovies"),
time = "ASAP")The function above calls a pizza place and provides several arguments (your address, pizza toppings, and delivery time). With some luck, the function will sucessfully return a new object (your cat’s favorite pizza). Note that when you have more than one argument, you will use a comma to separate them.
Mean age
We already covered two functions: c() and data.frame(). Now let’s use the mean() function to find the average age of our cats.
# Call the mean function with cat_ages as input
cat_ages_mean <- mean(cat_ages) # Assigns the output to cat_ages_mean
# Print the cat_ages_mean value to the console
cat_ages_mean## [1] 8.166667The mean() function takes the cat_ages vector as input, performs some calculations, and returns a single numeric object. Note that we assigned the output object to the name cat_ages_mean. If you don’t assign the output object it will be printed to the console and won’t be saved. Sometimes this is okay, especially when you’re still in the exploratory stages.
# Alternative without assigning output
mean(cat_ages) ## [1] 8.166667Note that our cat_ages vector has not changed at all. Each function has its own “environment”, and all of its calculations happen inside its own bubble. Usually anything that happens inside a function won’t change objects outside of the function’s environment.
cat_ages## [1] 3.0 7.0 14.5Function summary
There are functions in R that are more complex, but most boil down to the same general setup:
new_output <- function(input1, input2)
You call the function with input arguments inside parentheses and get an output object in return. You can make your own functions in R and call them almost anything you like, even my_amazing_cat_function(). Naming rules for functions and arguments are the same as those for objects (no spaces or special characters like +, -, *, /, \, =, or ! and they can’t begin with a number, sorry).
Pop Quiz!
Which of these is a valid function call?
lick(“paws” “tail”)
scratch, “couch”, “door”
sleep(3, “hours”)
meow(until fed)
shed(1 million, “hairs”)
Show solution
sleep(3, "hours")
Correct! You’re quite good at this.
3 | Reading and loading data
The first step of a good mystery is finding some clues. Here’s an example data table showing my favorite cats. It is saved on the internet as an Excel file here.
| Cat Name | Cat Age | Cat Color |
|---|---|---|
| Mr. Sauce | 3 | Salt-n-pepper |
| Sad Face | 2 | Tabby |
| Noodles | 6 | Calico |
But how do we get this data into R?
3.1 CSV to the rescue
The main data format in R is the CSV (comma-separated values). A CSV is a simple text file that can be opened in R and most other stats software, including Excel.
Here’s how the example cat table looks when it is saved as a .CSV file.
my_cats.csv
Cat Name,Cat Age,Cat Color
Mr. Sauce,3,Salt-n-pepper
Two Face,2,Tabby
Noodles,6,Calico It looks squished together right now, but that’s okay. When it’s opened in R the text will become a familiar looking table with columns and rows.
3.2 Save Excel to CSV file
First, open the Excel cat table by copying this path into a new window or to the Windows search bar - X:\Agency_Files\Outcomes\Risk_Eval_Air_Mod\_Air_Risk_Evaluation\R\R_Camp\Student Folder\my_cats.xlsx.
And then follow these instructions to save the Excel file as a CSV file.
- Go to File
- Save As
- Browse to your project folder
- Create new “data” folder
- Save as type: _CSV (Comma Delimited) (*.csv)_
- Any of the CSV options will work
- Any of the CSV options will work
- Click Yes
- Close Excel (Click “Don’t Save”)
Now let’s check that it worked. Return to RStudio and open your new data folder by finding it in your Files tab in the lower right window. Click on your CSV file and choose View File.
3.3 Read CSV into R
Copy the code below to your R script and run the line with the read.csv function (Hit CTRL + ENTER). It will return a nice cat data frame called my_cats. The character string "data/my_cats.csv" inside the parentheses of the read.csv() function is the path of the data file.
read.csv("data/my_cats.csv")## Cat.Name Cat.Age Cat.Color
## 1 Mr. Sauce 3 Salt-n-pepper
## 2 Sad Face 2 Tabby
## 3 Noodles 6 CalicoNote: The location of the CSV file above will only work if you have your project open. When your project is open, R sets the working directory automatically to your project folder. We will demonstrate reading a file directly from the X-drive a bit later.
3.4 Name your table
If you want to work with the data in R, you will need to give it a name by using the assignment operator <-.
Try the code below.
my_cat_file <- "data/my_cats.csv"
my_cats <- read.csv(my_cat_file)
# Type the name of the table to view it in the console
my_cats## Cat.Name Cat.Age Cat.Color
## 1 Mr. Sauce 3 Salt-n-pepper
## 2 Sad Face 2 Tabby
## 3 Noodles 6 Calico3.5 Bonus
You can save the file path as an object, such as cat_file <- "data/my_cats.csv". Then you can use that object as a shortcut to the location of your data. Now when you want to load the cat table you can write read.csv(cat_file). This handy trick will make it easier down the road when you want to update your code to use with new data.
# Assign the file path character string as an object with the name cat_file.
cat_file <- "data/my_cats.csv"
# Use read.csv with the object cat_file which refers to "data/my_cats.csv"
read.csv(cat_file)## Cat.Name Cat.Age Cat.Color
## 1 Mr. Sauce 3 Salt-n-pepper
## 2 Sad Face 2 Tabby
## 3 Noodles 6 Calico4 | Add a new package 📦
What is a package?
A package is a small add-on for R, like a phone App for your phone. They add capabilities like statistical functions, mapping powers, and special charts. In order to use a new package we first need to install it.
4.1 readr

The readr package helps import data into R in different formats. It does extra work for you like cleaning the data of extra white space and formatting tricky dates. Your packages are stored in your R library.
Add a package to your library
- Open RStudio
- Type
install.packages("readr")in the lower left console - Press Enter
- Wait two seconds
- Open the
Packagestab in the lower right window of RStudio to see the packages in your library- Use the search bar to find the
readrpackage
- Use the search bar to find the
The packages tab only shows the available packges that are installed. To use one of them, you will need to load it. Loading a package is like opening an App on your phone. To load a package we need to use the library() function. After loading the readr package you will able to read the cat data with the shiny new function read_csv(). This function is 300% better than read.csv().
library("readr")
my_cat_file <- "data/my_cats.csv"
read_csv(my_cat_file)## Parsed with column specification:
## cols(
## `Cat Name` = col_character(),
## `Cat Age` = col_integer(),
## `Cat Color` = col_character()
## )## # A tibble: 3 x 3
## `Cat Name` `Cat Age` `Cat Color`
## <chr> <int> <chr>
## 1 Mr. Sauce 3 Salt-n-pepper
## 2 Sad Face 2 Tabby
## 3 Noodles 6 CalicoPro-tip!
You may have noticed the row of three letter abbreviations under the column names. These describe the data type of each column.
chrstands for character vector, or a string of characters. Examples: “apple”, “apple5”, “5 red apples”
intstands for integer. Examples: 5, 34, 1071
We’ll see more data types, such as dates and logical, in later lessons.
Pop Quiz!
What data type is the Color column?
letters
character
words
numbers
integer
Show solution
character
## Colors cannot be applied in this environment :( Try using a terminal or RStudio.##
## --------------
## Meow! Not too shabby for a tabby.
## --------------
## \
## \
## \
## |\___/|
## ) (
## =\ /=
## )===(
## / \
## | |
## / \
## \ /
## jgs \__ _/
## ( (
## ) )
## (_(
## 4.1.1 Get help on functions
Let’s look a bit closer at the read_csv() function.
read_csv(my_cat_file)
# Get help
?read_csvFunction arguments
For read_csv(), the character object argument my_cat_file is what the function uses to know where to find the data file to read. Funcitons often have more than one argument. Type ?read_csv into your console to see help in the lower-right pane that describes all of the function’s arguments and what they do. Many of the options have default arguments (such as col_names = TRUE), which the function will use if you don’t provide an alternative argument. A short scroll down in the help window will show you more details about the arguments and the values they take.
`function(arg1 = input1, arg2 = input2, arg3…)
The file argument tells us that the function expects a path to a file. It can be many types of files, even a ZIP file. Below that, you’ll see the col_names argument. This argument takes either TRUE, FALSE, or a character vector of column names. The default is TRUE, which means the first row in the CSV is used as the column names for your data.
Don’t like the column names? We can give new column names to the col_names argument like this:
my_cat_file <- "data/my_cats.csv"
#Assign desired column names as a character vector named column_names
column_names <- c("Name", "Age", "Color")
read_csv(my_cat_file, column_names)## # A tibble: 4 x 3
## Name Age Color
## <chr> <chr> <chr>
## 1 Cat Name Cat Age Cat Color
## 2 Mr. Sauce 3 Salt-n-pepper
## 3 Sad Face 2 Tabby
## 4 Noodles 6 CalicoWe now have the column names we want, but now the original column names in our CSV file show up as a row in our data. We want read_csv to ignore the first row. Let’s look through the help window and try to find an argument that can help us. The skip argument looks like it could be helpful. Sure enough, the description is exactly what we’re looking for here. The default is skip = 0 (read every line), but we can skip the first line by providing skip = 1.
column_names <- c("Name", "Age", "Color")
read_csv(my_cat_file, column_names, skip = 1)## Parsed with column specification:
## cols(
## Name = col_character(),
## Age = col_integer(),
## Color = col_character()
## )## # A tibble: 3 x 3
## Name Age Color
## <chr> <int> <chr>
## 1 Mr. Sauce 3 Salt-n-pepper
## 2 Sad Face 2 Tabby
## 3 Noodles 6 CalicoSuccess!
You may be wondering why we included skip = for the skip argument, but only provided the objects for the other two arguments. When you pass inputs to a function, R will assume you’ve entered them in the same order that is shown on the ?help page. Let’s say you had a function called feed_pets() with 3 arguments:
feed_pets(dogs = "dogfood", cats = "catnip", fish = "pellets").
A shorthand way to write this would be feed_pets("dogfood", "catnip", "pellets"). If we write feed_pets("dogfood", "pellets", "catnip"), the function will send fish pellets to your cat and catnip to your fish. No good. If you really wanted to write “pellets” second, you would need to tell R which food item belongs to each animal, such as feed_pets("dogfood", fish = "pellets", cats = "catnip").
The same thing goes for read_csv(). In read_csv(my_cat_file, column_names, skip = 1), R assumes the file is my_cat_file and that the col_names should be set to column_names. The skip = argument has to be included explicitly because skip is the 10th argument in read_csv(). If we don’t include skip =, R will assume the value we entered is meant for the function’s 3rd argument.
Pro-tip!
A handy shortcut to see the arguments of a function is to enter the name of the function in the console and the first parenthesis, such as read_csv(, and then hit TAB on the keyboard. This will bring up a drop-down menu of all the available arguments for that function.
Key terms
package An add-on for R that contains new functions someone created to help you. It’s like an App for R.
library The name of the folder that stores all your packages, and the function used to load a package.
Pop Quiz!
What package does read_csv() come from?
dinosaur
get_data
readr
dplyr
tidyr
Show solution
readr
Great job! You’re fur real.
How would you load the package catfinder?
catfinder()
library(“catfinder”)
load(“catfinder”)
package(“catfinder”)
Show solution
library("catfinder")
Excellent! Keep the streak going.
Data exploration
5 | Pet detective
5.1 The case of the mopey cat

On your way home from work you find a wet and mopey cat sitting on your front stoop. Oh my! It looks so sad, maybe you can bring it inside and give it a treat.
When you pick up the cat you notice a collar with a number on it.
HINT: Your cat’s tag number is the same as the last digit of your birthday. Weird coincidence right?
This gives you an idea! Your friend recently mentioned a list that has all the missing cats people have reported in the city. Maybe you can use the tag number to help find the lost kitty’s home.
5.2 It’s inspector time!

Let’s get our script ready for some detective work. Since we bill by the hour we’re going to be very thorough with our documentation.
Script comments
Add a brief description of your new case to the top of the script using the comment symbol #.
# This file documents my search for the home of a lost cat I found this evening.
# What I know so far
animal <- "cat"
tag_num <- 444
pet_detective <- "Agent Cooper"
# Next steps
# 1. Find the missing cat database.
# 2. Load the cat data.
#Now let’s find ourselves some clues.
6 | All the cats!
Thanks to your friend, you can download a list of all the missing cats in your town. Follow the steps below to read the cat data into R.
Find the file’s location
Open this URL in your browser https://github.com/MPCA-air/RCamp/tree/master/data, and find the file missing_cat_list.csv. To download, Right click on your cat’s file and select Save Link As…. Navigate to your project folder and save the file into a folder named data\. Don’t have a data folder? Go ahead and create a new one.
Add the file location to your script
Now you can paste the file’s location into the read_csv() function. Here’s a code snippet to get you started.
library("readr")
# Replace the `...` with the name of the file
all_cats_file <- "data/..."
# Replace the `...` with 'all_cats_file'
all_cats <- read_csv(...)Show solution
library("readr")
all_cats_file <- "X:/Agency_Files/Outcomes/Risk_Eval_Air_Mod/_Air_Risk_Evaluation/R/R_Camp/Student Folder/missing_cat_list.csv"
all_cats <- read_csv(all_cats_file)## Parsed with column specification:
## cols(
## name = col_character(),
## color = col_character(),
## age = col_integer(),
## gender = col_character(),
## country = col_character(),
## grumpy = col_integer(),
## fearful_of_people = col_integer(),
## playful = col_integer(),
## friendly_to_people = col_integer(),
## clumsy = col_integer(),
## greedy = col_integer(),
## owner_phone = col_character()
## )Copy a file’s location
In Windows there’s a handy trick to copy the path to a file on your computer:
- Hold Shift + Right click on the file name. Select “Copy as path” from the menu.
Pro-tip!
File paths in R use forward slashes (
/). In Windows you’ll need to switch backslashes (\) to forward slashes (/). A file on your desktop located atC:\Desktop\file.csvwould be read into R asread_csv("C:/Desktop/file.csv")One trick to quickly fix a long path name is to press CTRL + F. Then you can use search tool to find any (
\) and replace it with (/).
NOTE: It’s good practice to load the packages you will need at the top of your script. You will need to run these lines every time you open R or switch projects. If you forget, you’re likely to see this error message.
read_csv(my_cat_file)## Error in read_csv(my_cat_file): could not find function "read_csv"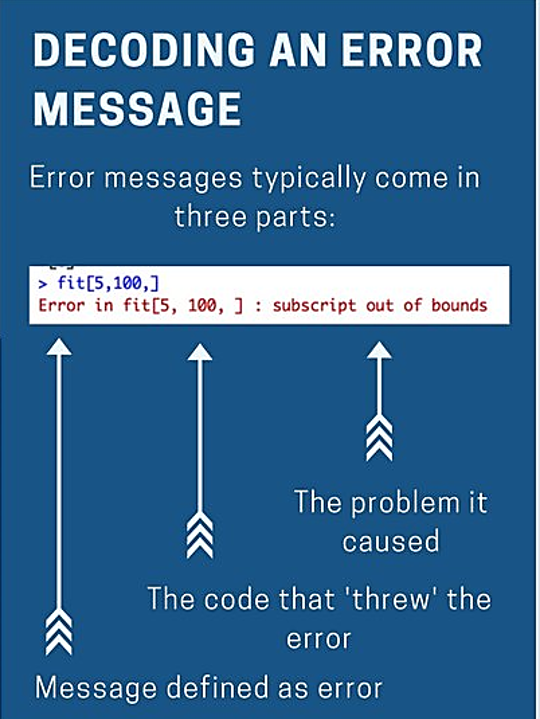
View the cats
Look in the upper right hand window of RStudio. This is the Environment window that shows all of the data frames you have created this session. You can see the names of the data frames, the number of observations (rows), and the number of variables (columns). This is helpful if you ever need to count your data (Hint Hint).
To see all the cat data click on the table called all_cats in the Environment window.
“There’s over 2,000 missing cats!”
That’s a lot of cats. And there’s more bad news. There’s no column for tag numbers. So which cat is yours? Let’s see… Does your cat look more like a Mr. Buttons or a Furry Potter? That’s a tough call. We need more information!
Let’s explore the missing cat data a bit more. Hopefully we can find some way to pick out your cat.
Pop Quiz, hotshot!
What is the second column in the all_cats data frame?
age
gender
nametag
color
fur
Show solution
color
You sure aren’t kitten around! Great work!
7 | dplyr

You’ve unlocked a new package!
The dplyr package is the go-to tool for exploring, re-arranging, and summarizing data.
Use install.packages("dplyr") to add dplyr to your detective library.
Quick stretch break
Stand up. Move around.
Your analysis toolbox: The key dplyr functions
| Function | Returns |
|---|---|
select() |
select individual columns to drop, keep, or reorder |
arrange() |
reorder or sort rows by value of a column |
mutate() |
Add new columns or update existing columns |
filter() |
pick a subset of rows by the value of a column |
group_by() |
split data into groups by values in a column |
summarize() |
calculate a single summary row for the entire table |
8 | select()
Use the select() function to drop a column you no longer need, to select a few columns to create a new sub-table, or rearrange the order of your table’s columns.
Drop a single column with a minus sign
library("dplyr")
# Drop the grumpy column
select(all_cats, -grumpy)## # A tibble: 2,785 x 11
## name color age gender country fearful_of_peop~ playful
## <chr> <chr> <int> <chr> <chr> <int> <int>
## 1 Latte grey and brown 3 Male Austral~ 2 5
## 2 Othello butterscotch 15 Female Thailand 1 7
## 3 Hershey tabby 5 Male Peru 1 6
## 4 Aboulomri brown 3 Female UK 1 2
## 5 Scratches white 11 Male Canada 1 5
## 6 Tuna salt and pepp~ NA Male Germany 1 7
## 7 Zonker white 4 Female China 5 5
## 8 Ovidius brown 10 Male Spain 2 5
## 9 Delia tabby 2 Female Canada 4 5
## 10 Hotdog salt and pepp~ 8 Female Spain 4 6
## # ... with 2,775 more rows, and 4 more variables:
## # friendly_to_people <int>, clumsy <int>, greedy <int>,
## # owner_phone <chr>Drop multiple columns with -c(col_1, col_2)
# Drop the grumpy and greedy columns
select(all_cats, -c(grumpy, greedy))## # A tibble: 2,785 x 10
## name color age gender country fearful_of_peop~ playful
## <chr> <chr> <int> <chr> <chr> <int> <int>
## 1 Latte grey and brown 3 Male Austral~ 2 5
## 2 Othello butterscotch 15 Female Thailand 1 7
## 3 Hershey tabby 5 Male Peru 1 6
## 4 Aboulomri brown 3 Female UK 1 2
## 5 Scratches white 11 Male Canada 1 5
## 6 Tuna salt and pepp~ NA Male Germany 1 7
## 7 Zonker white 4 Female China 5 5
## 8 Ovidius brown 10 Male Spain 2 5
## 9 Delia tabby 2 Female Canada 4 5
## 10 Hotdog salt and pepp~ 8 Female Spain 4 6
## # ... with 2,775 more rows, and 3 more variables:
## # friendly_to_people <int>, clumsy <int>, owner_phone <chr>Keep only three columns
# Keep the name, grumpy and greedy columns
select(all_cats, c(name, grumpy, greedy))## # A tibble: 2,785 x 3
## name grumpy greedy
## <chr> <int> <int>
## 1 Latte 2 2
## 2 Othello 2 2
## 3 Hershey 1 3
## 4 Aboulomri 7 7
## 5 Scratches 3 7
## 6 Tuna 1 2
## 7 Zonker 3 3
## 8 Ovidius 3 6
## 9 Delia 5 6
## 10 Hotdog 3 3
## # ... with 2,775 more rowsRearrange: Move the age and country columns directly after name
# Move the `age` and `country` columns directly after `name`
# Leave `everything()` else in same order
select(all_cats, name, age, country, everything())## # A tibble: 2,785 x 12
## name age country color gender grumpy fearful_of_peop~ playful
## <chr> <int> <chr> <chr> <chr> <int> <int> <int>
## 1 Latte 3 Austral~ grey and~ Male 2 2 5
## 2 Othello 15 Thailand buttersc~ Female 2 1 7
## 3 Hershey 5 Peru tabby Male 1 1 6
## 4 Aboulo~ 3 UK brown Female 7 1 2
## 5 Scratc~ 11 Canada white Male 3 1 5
## 6 Tuna NA Germany salt and~ Male 1 1 7
## 7 Zonker 4 China white Female 3 5 5
## 8 Ovidius 10 Spain brown Male 3 2 5
## 9 Delia 2 Canada tabby Female 5 4 5
## 10 Hotdog 8 Spain salt and~ Female 3 4 6
## # ... with 2,775 more rows, and 4 more variables:
## # friendly_to_people <int>, clumsy <int>, greedy <int>,
## # owner_phone <chr>9 | arrange()
Use the arrange() function to sort data based on one or more of the columns in the table. Let’s use arrange() to answer some questions about the missing cats.
- Which cats are the most grumpy?
- Which cats are the oldest?
- Are there cats from Australia?
- What are the oldest cats from Australia?
library("dplyr")
# Sort by the grumpy column
all_cats <- arrange(all_cats, grumpy)
# Enter the table name to view the ordered data
all_cats## # A tibble: 2,785 x 12
## name color age gender country grumpy fearful_of_peop~ playful
## <chr> <chr> <int> <chr> <chr> <int> <int> <int>
## 1 Hershey tabby 5 Male Peru 1 1 6
## 2 Tuna salt an~ NA Male Germany 1 1 7
## 3 Ludwig white 15 Male Peru 1 3 6
## 4 Beastie peaches~ 13 Male Thailand 1 2 5
## 5 Hayley grey an~ 1 Female USA 1 6 7
## 6 Fuzzina~ peaches~ 5 Male New Zea~ 1 2 6
## 7 Desiree black 9 Male Thailand 1 1 7
## 8 Zoloft butters~ 20 Female UK 1 2 7
## 9 Dinky salt an~ 4 Male Peru 1 1 7
## 10 Butters~ brown 6 Male Thailand 1 1 7
## # ... with 2,775 more rows, and 4 more variables:
## # friendly_to_people <int>, clumsy <int>, greedy <int>,
## # owner_phone <chr># To sort in descending order (highest to lowest)
# Add desc() around the column name
all_cats <- arrange(all_cats, desc(grumpy))
# View the top 5 rows using head()
head(all_cats)## # A tibble: 6 x 12
## name color age gender country grumpy fearful_of_peop~ playful
## <chr> <chr> <int> <chr> <chr> <int> <int> <int>
## 1 Aboulo~ brown 3 Female UK 7 1 2
## 2 Abigail grey and b~ 5 Male Canada 7 3 7
## 3 Broody brown 17 Female China 7 1 7
## 4 Albert butterscot~ 15 Male Canada 7 7 1
## 5 Barbra striped 11 Female Thaila~ 7 1 6
## 6 Alfred peaches an~ 14 Male Peru 7 7 5
## # ... with 4 more variables: friendly_to_people <int>, clumsy <int>,
## # greedy <int>, owner_phone <chr># Find the Aussie cats
# Find the ancient Aussie catsPro-tip!
When you save an arranged data table it maintains its order. This is perfect for sending people a quick Top 10 list of pollutants or sites.
10 | filter()
The filter() function creates a subset of the data based on the value of one or more columns. Use filter() to answer the questions below.
What are the cat names that are only 1 year old?
filter(all_cats, age == 1)Pro-tip!
We use a
==(double equals sign) for comparing values. A==makes the comparison “is it equal to?” and returns a True or False answer. So the code above returns all the rows where the conditionage == 1is TRUE.A single equals sign (
=) is used within functions to set options, for exampleread_csv(file = "mydata.csv"). Don’t worry too much. If you use the wrong symbol R is often helpful and will let you know which one is needed.
10.1 Comparisons
To use filtering effectively you’ll want to know how to select observations using various comparison operators.
Key comparison operators
| Symbol | Comparison |
|---|---|
> |
greater than |
>= |
greater than or equal to |
< |
less than |
<= |
less than or equal to |
== |
equal to |
!= |
not equal to |
%in% |
value is in a list |
What are the cat names that have a grumpy score greater than 6?
filter(all_cats, grumpy ...)What are the cat names that are either striped or orange in color?
filter(all_cats, color %in% c("orange", "striped"))What is that
c()thing all about again?
You can put multiple values inside c() to make a vector of items. Each item in the vector is separated by a comma. Let’s create a short vector of your favorite colors.
# This is an example vector
my_fave_colors <- c("green", "orange", "cornflower")
my_fave_colors## [1] "green" "orange" "cornflower"10.2 Multiple filters
Exercise
Create a small table called uk_oldies that only has cats from the UK and are older than 20.
Work with your neighbor to create the table.
Pop Quiz, hotshot!
Are there more cats with the color striped or butterscotch?
more butterscotch
more striped
same amount in both groups
Hint: Start by creating a new table called striped using the code filter(all_cats, color == ...)
Show solution
more striped
Feline fine after that exercise! Great job!
10.3 Cat challenge 🐱
Ask your neighbor a question about the cat data that requires the %in% operator.
Try to answer your neighbor’s question.
10.4 Cat challenge #2 🐱 🐱
Team up with your neighbor to think of a summary question to ask the table next to you about the missing cat data. Ask them the question and they’ll ask you a question about the cats in return.
Work with your neighbor to find the answer.
10.4.1 Play time
Now that you are more familiar with the data, maybe you can use the personality traits of the missing cats to find which one is yours.
If only you knew the personality of your cat.
Spend some time getting to know your cat. Is it already hiding from you under the couch? Does it seem Australian? Is it a young cat? Once you’ve gotten to know your cat, move on to the next section.
11 | Lost cat traits
Congratulations! You’ve unlocked your cat’s personality.
Thanks to all the quality time you spent with your cat, you were able to collect some high quality data describing your cat’s personality. You can use your cat’s tag number to find the CSV file for your cat. Download your file from the Cat personality files.
To download, Right click on your cat’s file and select Save Link As…. Navigate to your project folder and save the file into a folder named data\.
Now you can read in the file with the personality traits of your cat.
library("readr")
# Replace the `##` with your tag number
my_cat <- read_csv("data/foundcat_tag##.csv")Take a minute to explore your cat’s personality traits. Can you guess its name?
11.1 Find your cat
Almost there! Now you can use filter() to check if the all_cats table has a cat with a similar personality as yours. Let’s hope so.
Use filter() to find your cat in the big all_cats table.
# Replace the `?` marks with your cat's personality traits
the_one <- filter(all_cats,
greedy == ?,
playful == ?,
age ...)Hint: You can also use the cat’s
agerange to help narrow down the number of potential cats.
Great work!
You found your cat a home and you’ve earned yourself a detective badge.

Meet the R cats
Let’s introduce our rescued cats.

Possible topics:
- Your found cat’s name and best personality trait.
- Your name or detective name.
- Something you want to learn to do with data?
- Are you a Cat person? Dog person? Lizard person? Fish person? Plant person?
RECAP:
What packages have we added to our
library?What new functions have we learned?
Survey says
On the front of your sticky note answer one of these:
- Something you really liked learning today?
- A useful thing you learned?
- A new skill you are you excited about using?
On the back:
- A lingering question you have about the material.
- A topic that was confusing and could use more clarification?
We will compile the questions and send out answers before next class. If you think of something later, please e-mail us any questions you have. If you’re uncertain about something I guarantee someone else is as well. So help a friend, and ask a question.
Shutdown complete
When you close RStudio it will ask you about saving your workspace and other files. This can get tiresome after awhile. Follow the steps below to set these options once and for always.
Turn off “Save Workspace”
- Go to Tools on the top RStudio navigation bar.
- Choose
Global Options.... - On the first screen:
- Set Save workspace to .RData on exit: to “Never”.
- Uncheck Always save history
- Uncheck Restore .Rdata into workspace at startup
Congratulations! You’ve completed day 1.
